Research
Mechanisms of gene regulation by RNA cleavage and decay machineries
We study several RNA cleaving and degrading machineries in human cells that are involved in gene regulation and RNA processing. Our goal is a comprehensive view of these intricate processes by combining the characterisation of well-defined reconstituted systems with studies in the complex environment of human cells. We exploit modern structural methods (crystallography, cryo-electron microscopy) together with quantitative biochemical analyses, as well as next-generation sequencing and proteomics tools.
Reserach Highlights
Nucleic acid binding module of Integrator complex
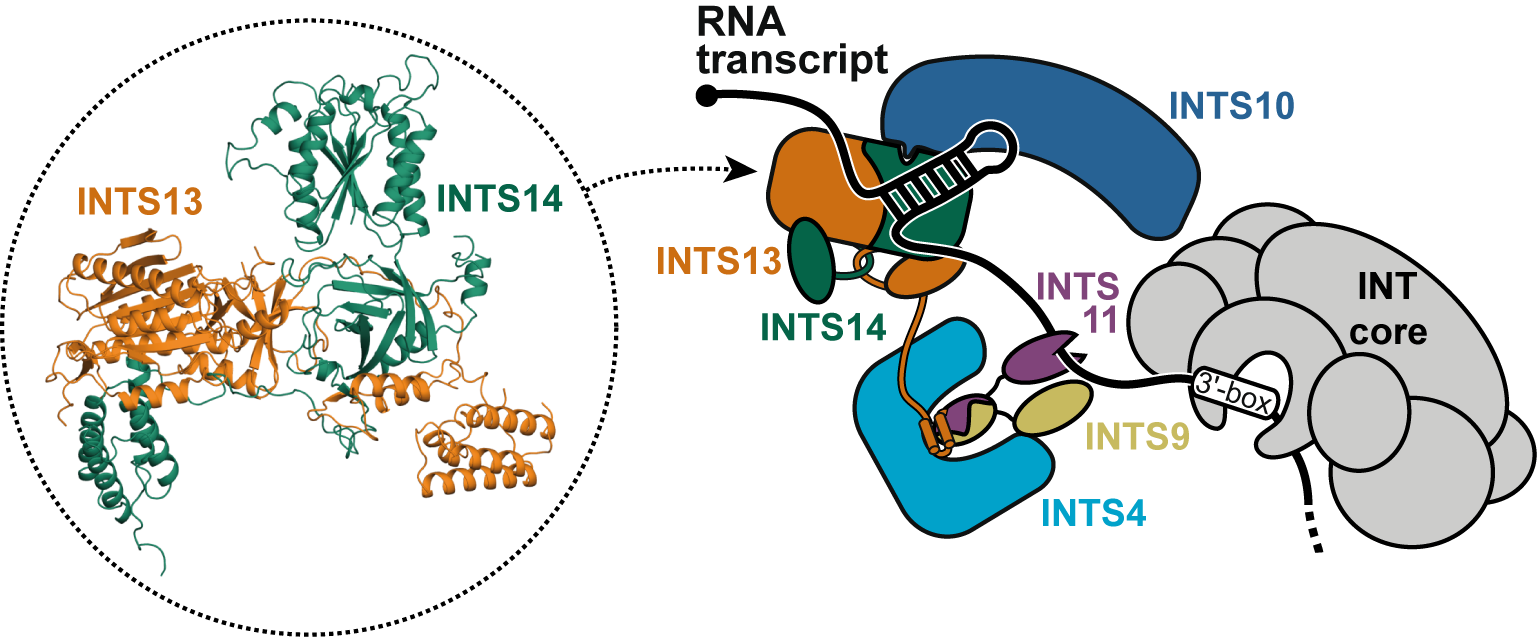
The Integrator complex is the 3'-end processing machinery for snRNAs and an attenuator for mRNA transcription. We identified a new module of the complex that binds nucleic acids and forms a direct connection to the RNA cleavage module. This connection is required for the module's accessory role in snRNA processing. The module binds preferentially to RNA hairpins and we could show that its subunits are important during transcription attenuation of mRNAs. Our work suggests that mutations and deletions in the nucleic acid binding module lead to defects in spermatogenesis or ciliogenesis because they impair proper recruitment of the RNA cleavage module to RNA transcripts.
Publication
Sabath K, Stäubli ML, Leitner A, Moes M, Jonas S#. INTS10–INTS13–INTS14 form a functional module of Integrator that binds nucleic acids and the cleavage module. 11, 3422 (2020) Nat. Commun. external page doi: 10.1038/s41467-020-17232-2
Activation of RNA Helicases in mRNA Splicing and Ribosome Biogenesis
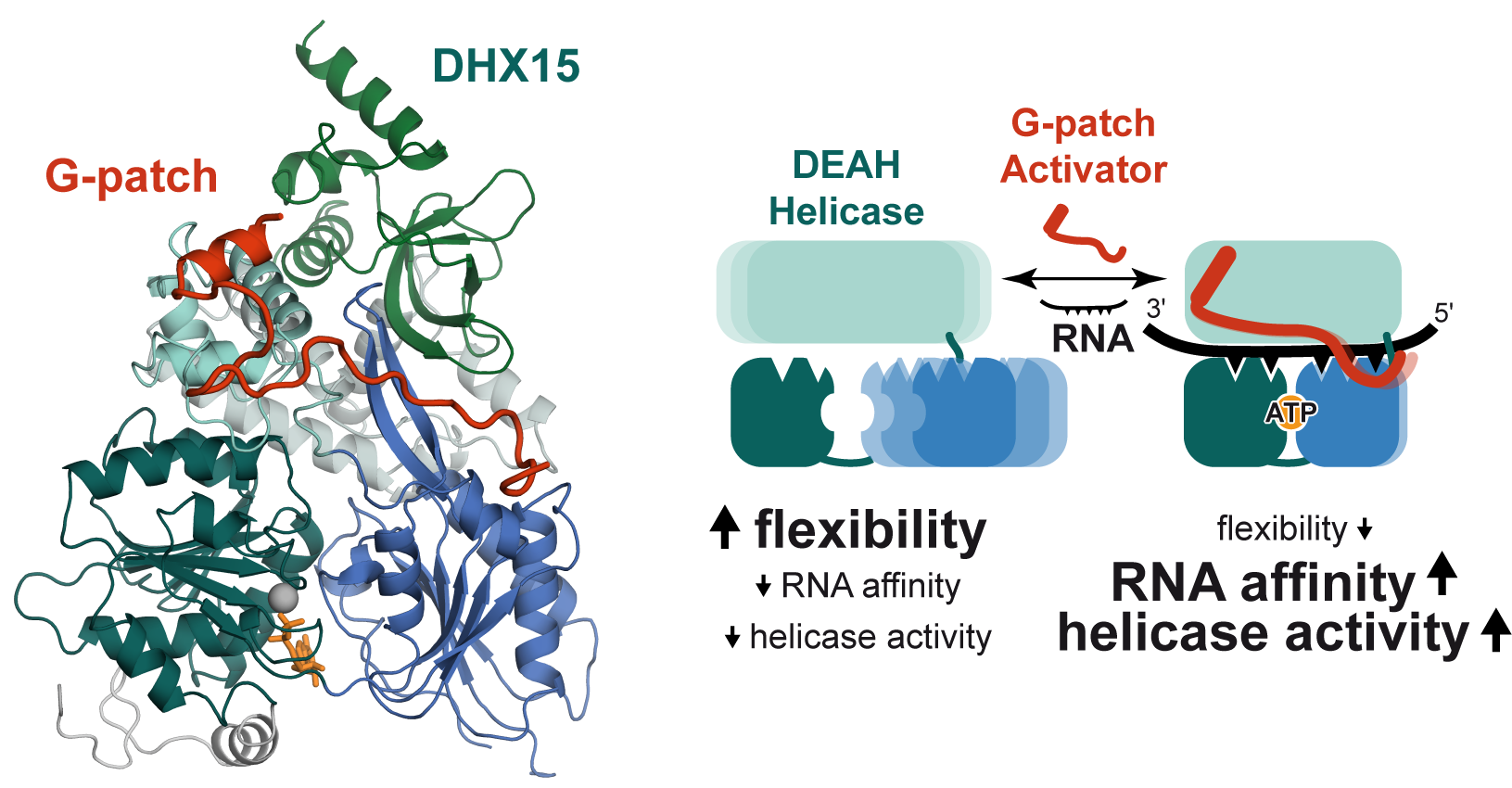
A family of RNA helicases, which is called DEAH helicases, has important tasks at pivotal points during splicing of mRNAs or assembly of ribosomes. DEAH helicases work by translocating along RNA and thereby unwind RNA duplexes or dissociate bound proteins. Because these RNA helicases are poor enzymes without intrinsic selectivity for their target RNAs, they require adapter proteins that recruit them to functional sites and simultaneously enhance their catalytic activity. One essential class of DEAH activators is formed by G-patch proteins, which bind helicases via their eponymous glycine-rich motif. We solved the structure of a G-patch bound to the human DEAH helicase DHX15, and characterized biochemically how it activates the helicase. Our analysis shows that G-patches tether mobile sections of DEAH helicases together and activate them by stabilizing a functional conformation with high RNA affinity.
Publication
Studer MK, Ivanović L, Weber ME, Marti S, Jonas S#, Structural basis for DEAH-helicase activation by G-patch proteins. (2020) Proc. Natl. Acad. Sci. USA. external page doi: 10.1073/pnas.1913880117
Previous Research
mRNA decay machineries
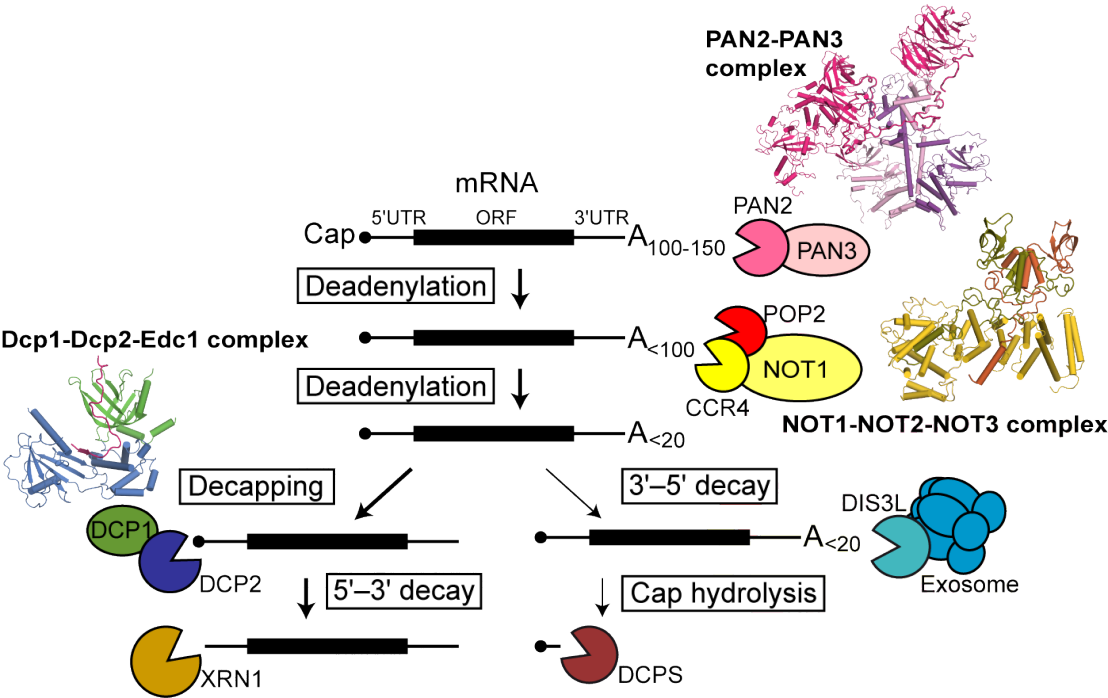
In eukaryotes mRNAs are degraded in a number of mRNA quality control pathways, and also during general turnover. 5'-3' mRNA decay proceeds via deadenylation followed by decapping, and finally rapid digestion by exonuclease Xrn1. Deadenylation is also involved in translational repression of mRNAs, which is of importance during the cell cycle, miRNA mediated gene silencing and embryonic development.
We illuminated structures and mechanisms of eukaryotic deadenylation and decapping complexes
- of the two major deadenylase complexes in eukaryotes, the CCR4-NOT and the PAN2-PAN3 complex
- as well as mechanism of activation of the decapping enzyme Dcp2 by its enhancers Dcp1, Edc1 and Edc4.
Selected Publications
Valkov E, Muthukumar S, Chang CT, Jonas S#, Weichenrieder O#, Izaurralde E#. Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation. 23, 574–579 (2016) Nat. Struct. Mol. Biol. doi: external page 10.1038/nsmb.3232
Jonas S*, Christie M*, Peter D, Bhandari D, Loh B, Huntzinger E, Weichenrieder O, Izaurralde E. An asymmetric PAN3 dimer recruits a single PAN2 exonuclease to mediate mRNA deadenylation and decay. 21, 599–608 (2014) Nat. Struct. Mol. Biol. doi: external page 10.1038/nsmb.2837
Chang CT, Bercovich N, Loh B, Jonas S, Izaurralde E. The activation of the decapping enzyme DCP2 by DCP1 occurs on the EDC4 scaffold and involves a conserved loop in DCP1. 42, 5217–5233 (2014) Nucleic Acids Res. doi: external page 10.1093/nar/gku129
Boland A*, Chen Y*, Raisch T*, Jonas S*, Kuzuoğlu-Öztürk D, Wohlbold L, Weichenrieder O, Izaurralde E. Structure and assembly of the NOT module of the human CCR4-NOT complex. 20, 1289–1297 (2013) Nat. Struct. Mol. Biol. doi: external page 10.1038/nsmb.2681
post-transcriptional gene regulation
Nonsense-mediated decay
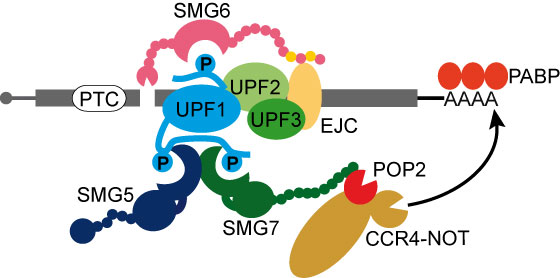
The nonsense-mediated mRNA decay (NMD) pathway triggers destruction of aberrant mRNAs with premature termination codons (PTC). In addition to its quality control function, NMD is also involved in post-transcriptional regulation of 5-10% of the human transcriptome.
We determined, how three NMD factors SMG5, SMG6 and SMG7 bring about efficient mRNA degradation after PTC recognition.
- SMG5 and SMG7 form a pseudosymmetric dimer via two phosphopeptide binding domains. Heterodimer formation is required for simultaneous binding of two phosphorylated sites of UPF1 within a defined spacing and with high affinity.
- SMG7 uses its unstructured C-terminus to directly recruit the general mRNA decay machinery to the target mRNA. It binds the processive deadenylation complex (CCR4-NOT) on one of its catalytic subunits.
- We identified two motifs in SMG6 that directly bind the exon-junction complex. These motifs provide a second contact point for SMG6 on the assembled NMD machinery in addition to phospho-UPF1, which results in both higher affinity and specificity of recognition.
Selected Publications
Jonas S, Weichenrieder O, Izaurralde E. An unusual arrangement of two 14-3-3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. 27, 211–225 (2013) Genes Dev. doi: external page 10.1101/gad.206672.112
Loh B, Jonas S, Izaurralde E. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. 27, 2125–2138 (2013) Genes Dev. doi: external page 10.1101/gad.226951.113
Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. 24, 2440–2450 (2010) Genes Dev. doi: external page 10.1101/gad.604610
Nanos-mediated gene regulation
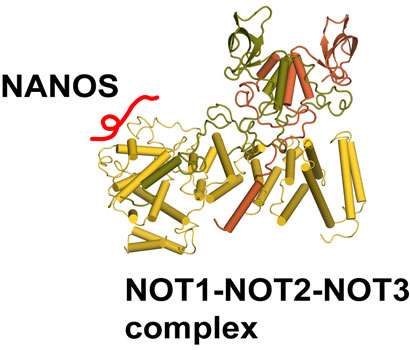
We elucidated how human Nanos orthologs, which are required for germ cell development, recruit the NOT-module of the CCR4-NOT deadenylase complex to mediate translational repression of mRNA-targets. Our findings provide the structural basis for binding of all vertebrate Nanos to NOT1, and identify the CCR4-NOT complex as the main effector complex for Nanos function.
Publication
Bhandari D, Raisch T, Weichenrieder O, Jonas S#, Izaurralde E#. Structural basis for the Nanos-mediated recruitment of the CCR4-NOT complex and translational repression. 28, 888–901 (2014) Genes Dev. doi: external page 10.1101/gad.237289.113